MAGERI: Molecular tAgged GEnome Re-sequencing pIpeline¶
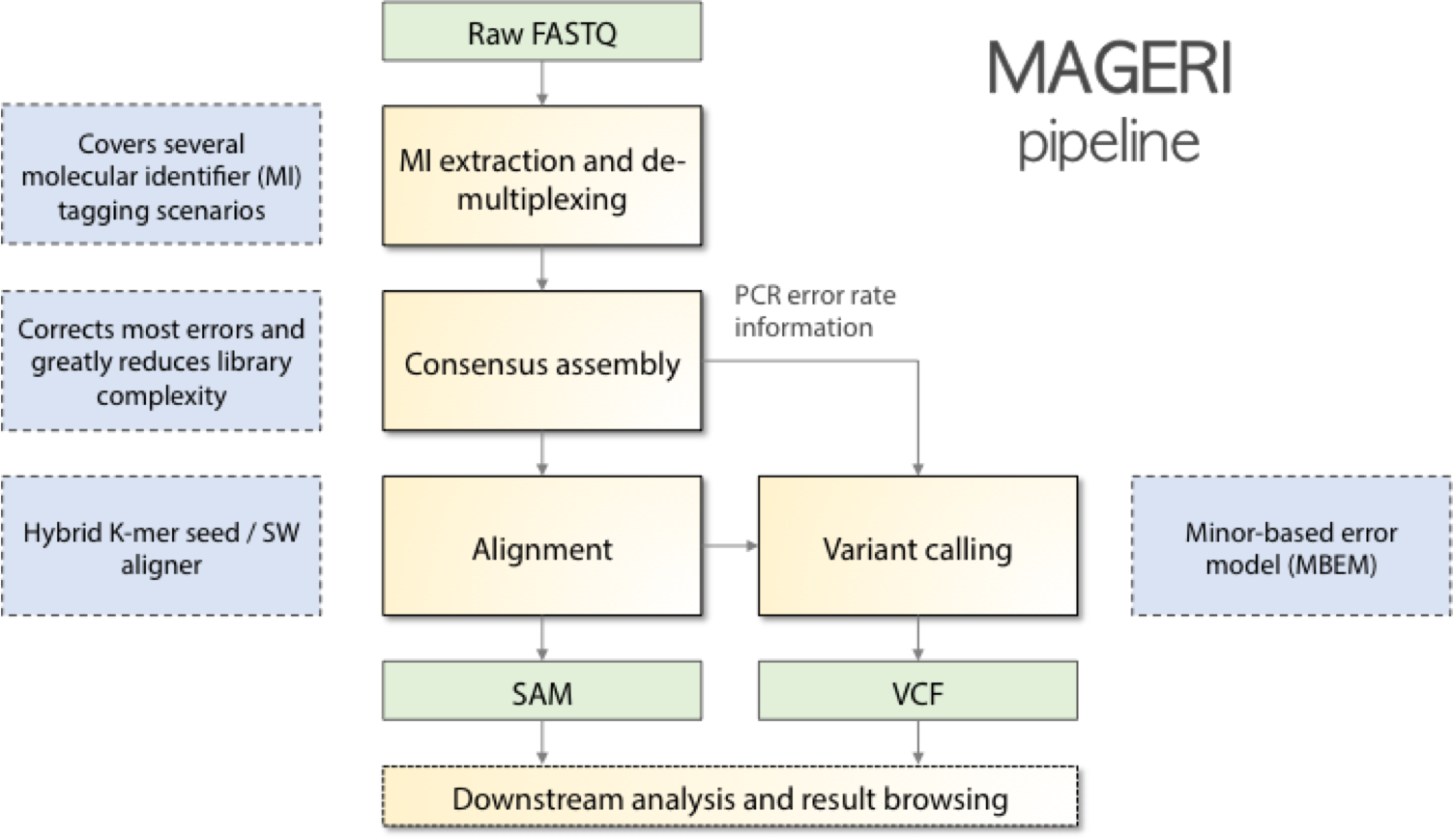
MAGERI is an all-in-one software for analysis of targeted genome re-sequencing data for libraries prepared with novel unique molecular identifier tagging technology. Starting from raw sequencing reads, MAGERI extracts UMI sequences, performes primer and adapter matching and trimming, assembles molecular consensuses, alignes them to reference sequences and calls variants. MAGERI output is provided in conventional SAM and VCF formats, so it can be browsed and post-processed by the majority of conventional bioinformatics software.
Terminology¶
- UMI - unique molecular identifier, a short (4-20bp) degenerate nucleotide sequence, that is attach to cDNA/DNA molecules in order to trace them throughout the entire experiment.
- Sample barcode - a short specific nucleotide sequence used to mark cDNA/DNA molecules that correspond to a given sample in a pooled sequencing library
- MIG - molecular identifier group, a set of reads or read pairs that have an identical UMI sequence
- MIG consensus - the consensus sequence of MIG, that is, the consensus of multiple alignment of all reads in a given MIG
- CQS - consensus quality score, calculated as the fraction of reads matching the consensus sequence at a given position. Can be scaled to
[2, 40]range to fit Phred33 quality representation. - Major variant (aka dominant variant, supermutant) - a sequence variant that is present in MIG consensus, but doesn’t match the reference sequence
- Minor variant - a sequence variant that differs from the consensus sequence found in one or more reads within a given MIG